
Extended Access feature | Programmatic access


|
Table of Contents
: Accessing data
: Extended Access feature | Programmatic access |

|
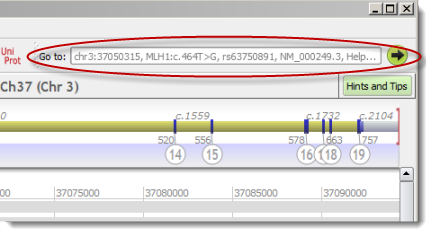
To complete the Focus on dialog (menu 'View' > 'Focus on...'), Alamut® Visual provides a new Extended Access Feature allowing:
Example queries:
| Query by reference | Comment |
|---|---|
MLH1 | Official HGNC gene symbol |
Breast cancer | Disease, previous symbol or gene alias |
HGNC:7127 | HGNC Id |
NM_000249.3 | cDNA RefSeq Id |
ENST00000231790 | Ensembl Transcript Id (mapped to a NCBI RefSeq) |
NG_007941.2 | RefSeqGene Id |
LRG_1 | LRG Id |
NP_000240.1 | Protein RefSeq Id |
P40692 | UniProt Id |
rs63750891 | Reference SNP Id |
COSM35380 | COSMIC Id |
OMIM:120436 | OMIM� Id |
Example queries:
| Query by position | Comment |
|---|---|
chr3:g.37050315 | Standard gDNA query (implicitly on GRCh37) |
3:37050315 | Short gDNA query |
3:36000000-38000000 | Interval gDNA query (response is limited to 50 genes) |
chr3(NCBI36):g.37025319 | Use given assembly |
hg18:3:37025319 | Use given assembly (alternative writing) |
g.37050315 | gDNA query inside current gene |
NM_000249.3:c.464 | cDNA position query |
NM_000249.3:464 | Short cDNA position query |
ENST00000231790:c.464 | cDNA position query on Ensembl Trancript Id (mapped to a NCBI RefSeq) |
NM_000249.3:p.155 | Protein position query |
NM_000249.3:p.Leu155Arg | Protein substitution (only position is used) |
NM_000249.3:L155R | Protein substitution short query (only position is used) |
P603R, K618R, Q689R | Substitution list, e.g. from literature (implicitly on current gene — only position is used) |
![]() In the above queries, a cDNA RefSeq can be replaced by a gene symbol (e.g.,
In the above queries, a cDNA RefSeq can be replaced by a gene symbol (e.g.,
MLH1:c.464) or a RSG/LRG ID
(e.g., LRG_1:c.359,
NG_007941.2:p.211).
![]() When a gene is already open in Alamut® Visual, neither RefSeq nor gene symbol may be given
(in that case, protein substitutions require the standard prefix, e.g.
When a gene is already open in Alamut® Visual, neither RefSeq nor gene symbol may be given
(in that case, protein substitutions require the standard prefix, e.g.
p.L155R).
![]() For gDNA queries, the default assembly is GRCh37.
For gDNA queries, the default assembly is GRCh37.
Example queries:
| Query by variant | Comment |
|---|---|
chr3:g.37050315T>G | Query with genomic variation (implicitly on GRCh37) |
3:37050315T>G | Short query with genomic variation |
chr3(NCBI36):g.37025319T>G | Use given assembly |
hg18:3:37025319T>G | Use given assembly (alternative writing) |
NM_000249.3:c.464T>G | cDNA variant |
ENST00000231790:c.464T>G | cDNA variant on Ensembl Trancript Id (mapped to a NCBI RefSeq) |
c.157_160delGAGG, c.2157dupT and c.-64G>T | Variant list, e.g. from literature (implicitly on current gene) |
![]() In the above cDNA query, RefSeq can be replaced by a gene symbol (e.g.,
In the above cDNA query, RefSeq can be replaced by a gene symbol (e.g.,
MLH1:c.464T>G) or a RSG/LRG ID
(e.g., LRG_1:c.359G>A).
![]() For gDNA variants, the default assembly is GRCh37.
For gDNA variants, the default assembly is GRCh37.
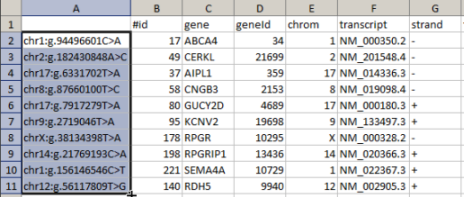
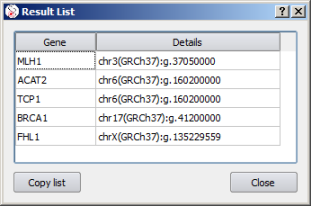
Several lines from an NGS output file can be copied and pasted into the input field:
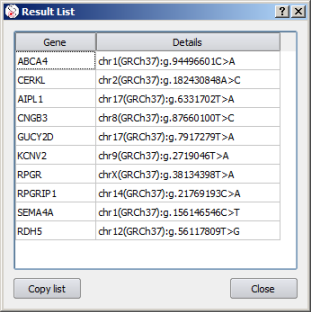
The pasted data then shows up in a variant list. Double-click on an entry to jump from a variant to another:
![]() Data can also be dragged and dropped in the entry field.
Data can also be dragged and dropped in the entry field.
![]() Lines from any BED file may also be pasted into the entry field. In that case, the only 2 first colums
(chromosomes,
gDNA start) will be taken into account.
Be aware that, as BED files use 0-based coordinates, the focus within Alamut® Visual will be
shifted by 1 nucleotide.
Lines from any BED file may also be pasted into the entry field. In that case, the only 2 first colums
(chromosomes,
gDNA start) will be taken into account.
Be aware that, as BED files use 0-based coordinates, the focus within Alamut® Visual will be
shifted by 1 nucleotide.
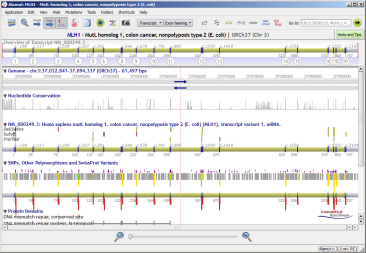
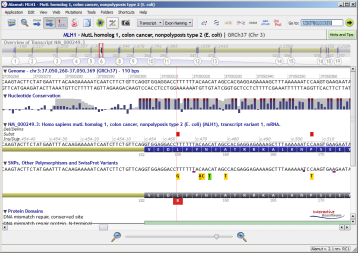
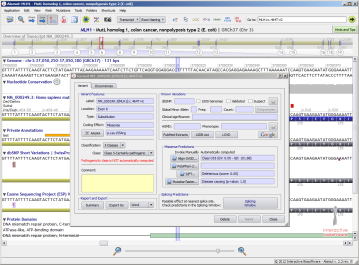
Depending on the query type, Alamut® Visual can provide different response types:
| Open gene | Direct focus to a location |
|---|---|
|  |
| Create variant | Position/Variant list |
 |
 |
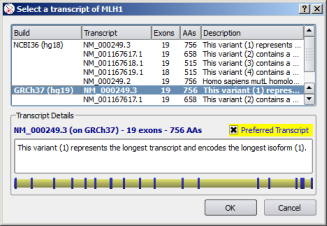
| Select gene transcript | Select gene |
 |
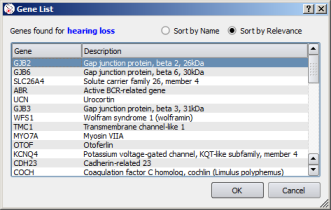 |
![]() For each gene result, the preferred assembly and/or transcript, if any, is automatically selected.
For each gene result, the preferred assembly and/or transcript, if any, is automatically selected.
Example queries (when a gene is already displayed):
| Query | Comment |
|---|---|
BED<http://www.interactive-biosoftware.com/alamut/doc/2.15/templates/MLH1.bed | Query with file format and URL |
GAAGGGAACCTGATTGGATTACCCCTTCTGATT | Query with nucleotide sequence |
BAM<http://www.your-institution.org/NGS/alignment.bam | Query with file format and URL |
© 2020 Interactive Biosoftware - Last modified: 30 December 2017