
Hints & tips | BENCHlab Integration | Menus


|
Table of Contents
: Miscellaneous
: Hints & tips | BENCHlab Integration | Menus |

|
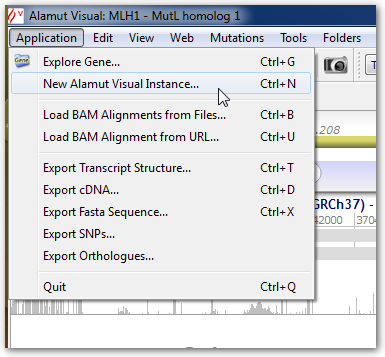
Several instances can be launched from the menu Application > New Alamut Visual Instance:
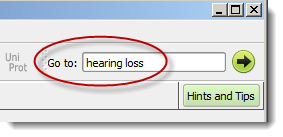
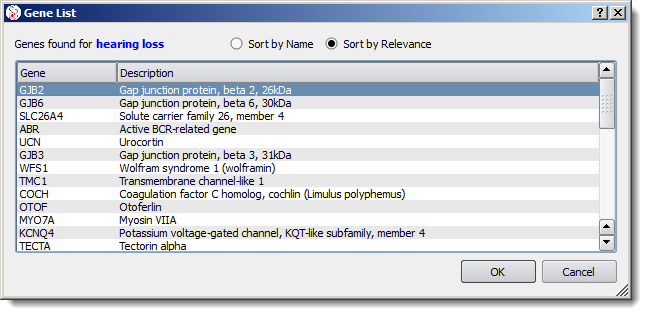
Type in a disease name or a phenotype to get a relevant gene list:
 |
 |
 |
![]() In July 2012, rare diseases from
orphadata
have been added to Interactive Biosoftware's Talamut engine,
see Orphadata web site: Diseases with their associated genes
managed by Orphanet.
Interactive Biosoftware is member of RD-Connect,
a recently funded EU project on: "Databases, biobanks and clinical bio-informatics hub for rare diseases".
In July 2012, rare diseases from
orphadata
have been added to Interactive Biosoftware's Talamut engine,
see Orphadata web site: Diseases with their associated genes
managed by Orphanet.
Interactive Biosoftware is member of RD-Connect,
a recently funded EU project on: "Databases, biobanks and clinical bio-informatics hub for rare diseases".
Transcript selection is directly available from the tool bar:
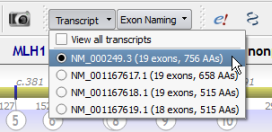
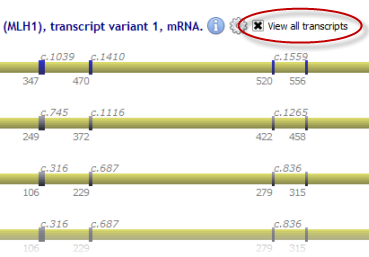
It is possible to display all available transcripts simultaneously:
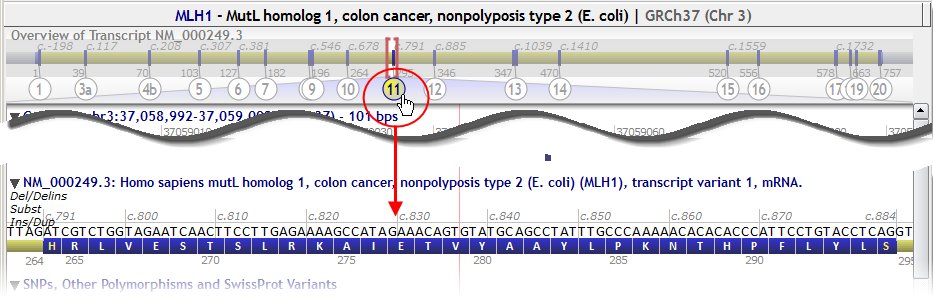
Click on an exon number in the gene overview to zoom in directly:
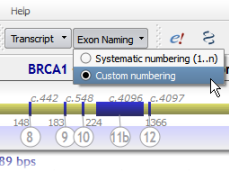
For genes built on the GRCh37 (hg19) assembly, we have used the NCBI RefSeq numbering scheme (along with simple systematic numbering).
However, feedback from a number of groups shows that legacy exon numbering schemes are still required. This is why we have restored these numbering schemes where appropriate (e.g., BRCA1, CFTR, NF1), as they already were in Alamut® Visual on NCBI36 (hg18).
Exon numbering selection is directly available from the tool bar:
It is also possible to select one of these 2 numberings in the Options dialog box (menu 'Tools' > 'Options' > 'Display' tab).
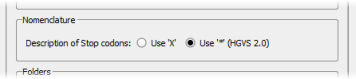
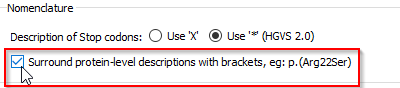
The HGVS 2.0 nomenclature uses '*' for Stop codons instead of 'X'. It is possible to select one of these 2 notations in the Options dialog box (menu 'Tools' > 'Options' > 'Preferences' tab).
The HGVS 15.11 nomenclature, applied by default in Alamut Visual 2.8, surrounds ( ) for the protein-level description. It is possible to unselect this default behaviour in the Options dialog box (menu 'Tools' > 'Options' > 'Preferences' tab).
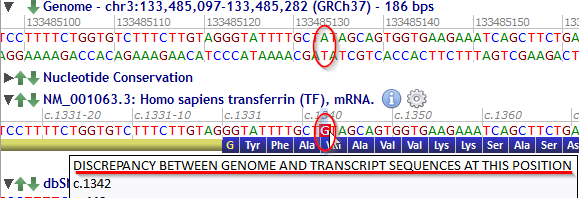
Transcript nucleotides are highlighted in red where they differ from the reference genome sequence and the tooltip gives a warning message.
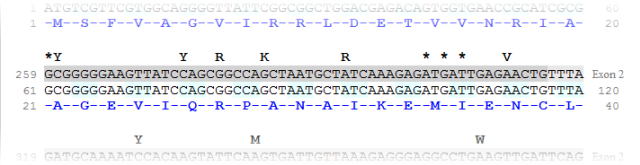
The menu 'Application' > 'Export cDNA' produces an HTML textual representation of the gene:
variations from dbSNP, exons, cDNA, codons and protein. Substitutions (from dbSNP) are represented using the IUPAC nomenclature
and other variations are represented by a '*' positionned on their 5' end.
![]() In Alamut® Visual, cDNA sequence export can include (intronic) exon flanking sequences, specified in number of bps.
In Alamut® Visual, cDNA sequence export can include (intronic) exon flanking sequences, specified in number of bps.
![]() No variation is represented if its 5' end is either located on a substitution or in an intronic region that is not displayed.
No variation is represented if its 5' end is either located on a substitution or in an intronic region that is not displayed.
![]() To import this HTML document into Word, use the menu 'Insertion' > 'File...'
from Word, then set the document font to 'Courier New' and adjust left and right margins.
To import this HTML document into Word, use the menu 'Insertion' > 'File...'
from Word, then set the document font to 'Courier New' and adjust left and right margins.
In the transcript track, just right-click on a nucleotide, press the shift key and maintain it while selecting all the nucleotides you want to copy, then move along the transcript with the mouse and select the last nucleotide wanted. Once done, simply right-lick on the selected sequence to copy and paste this one.
The cDNA sequence is copied to the clipboard.
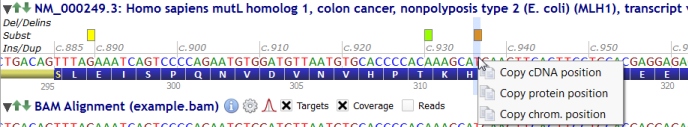
In the transcript track, just right-click on a nucleotide and select from the menu a position between cDNA, protein or genomic positions.
The position value is copied to the clipboard.
Use the following keys to navigate in Alamut® Visual:
Select 'Help' > 'Software Reference...' from the main menu of Alamut® Visual to copy its reference and paste it into your paper.
Visit the literature page on our web site.
© 2020 Interactive Biosoftware - Last modified: 30 December 2017