
Missense predictions | Splicing module | Micro-RNA predictions


|
Table of Contents
: In silico predictions
: Missense predictions | Splicing module | Micro-RNA predictions |

|
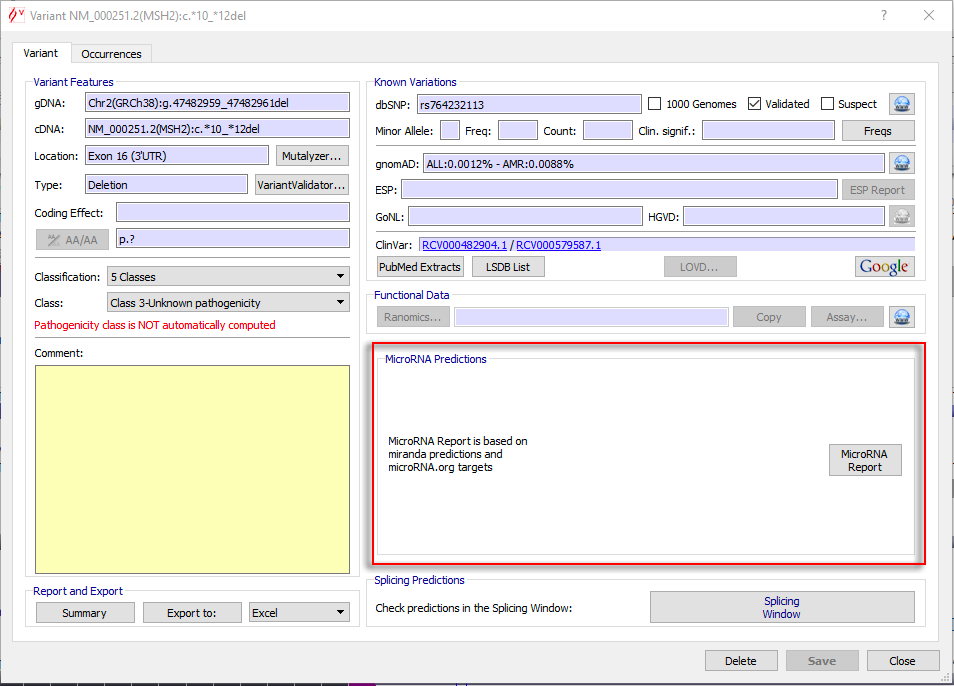
Alamut® Visual 2.1 introduced a new micro-RNA module. It enables the user to visualize 3'UTR variant impacts on micro-RNA (miRNA) target sites.
This module is available in the variant window when the variant is in the 3'UTR region.
This micro-RNA module relies on the public domain part of the miRanda algorithm (see microRNA.org).
This algorithm detects potential microRNA target sites in genomic sequences.
A dynamic programming local alignment is carried out between the query miRNA sequence (collected from the miRBase web site) and the genomic sequence.
This alignment procedure scores based on sequence complementarity and not on sequence identity
(the G:U wobble base pair is also permitted, but generally scores less than the more optimal matches).
Alamut® Visual points out micro-RNA target site prediction differences between the wild-type and the mutated sequences.
The miRanda parameters are set as: miranda -sc 120 -noenergy.
To launch the micro-RNA module, click on the 'MicroRNA Report' button from the variant window :
A micro-RNA report shows up after computation:
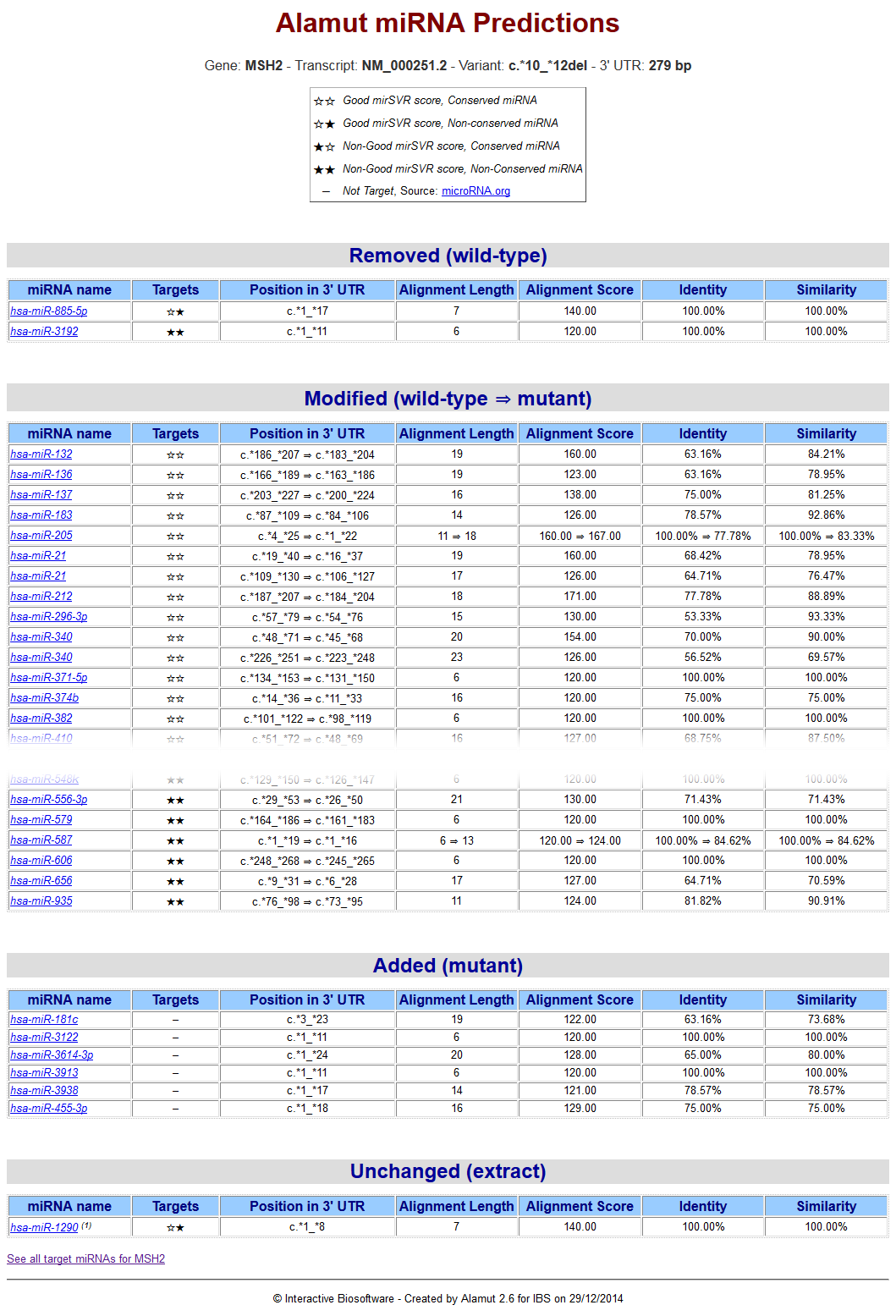
All micro-RNA predicted targets are classified in 4 sections: Removed, Modified, Added and Unchanged
for which 6 features are computed: targets, position in 3'UTR, alignment length, alignment score, identity and similarity.
An individual link to the miRBase description for each micro-RNA and a global link to the microRNA.org listing for all target micro-RNA are given.
Enright, AJ., John, B., Gaul, U., Tuschl, T., Sander, C., Marks, DS. (2003). MicroRNA targets in Drosophila. Genome Biology. 5 : R1.
John, B., Enright, AJ., Aravin, A., Tuschl, T., Sander, C., Marks, DS. (2004). Human MicroRNA targets. PLoS Biol, 3 : e264.
Betel, D., Wilson, M., Gabow, A., Marks, DS., Sander, C. (2007). The microRNA.org resource: targets and expression. Nucleic Acids Res. 36 : D149-53.
Betel, D., Koppal, A., Agius, P., Sander, C., Leslie, C. (2010). Comprehensive modeling of microRNA targets predicts functional non-conserved and non-canonical sites. Genome Biology 11 : R90
© 2020 Interactive Biosoftware - Last modified: 3 June 2019