
Missense predictions | Splicing module | Micro-RNA predictions


|
Table of Contents
: In silico predictions
: Missense predictions | Splicing module | Micro-RNA predictions |

|
Alamut® Visual 2.0 introduced automatically computed missense predictions, while still retaining the ability to query prediction sites manually, giving users the opportunity to adjust prediction settings and options.
Currently, automatically computed predictions are provided for Align GVGD, SIFT and MutationTaster.
Automatically computed predictions are available in the variant window and in variant export data.
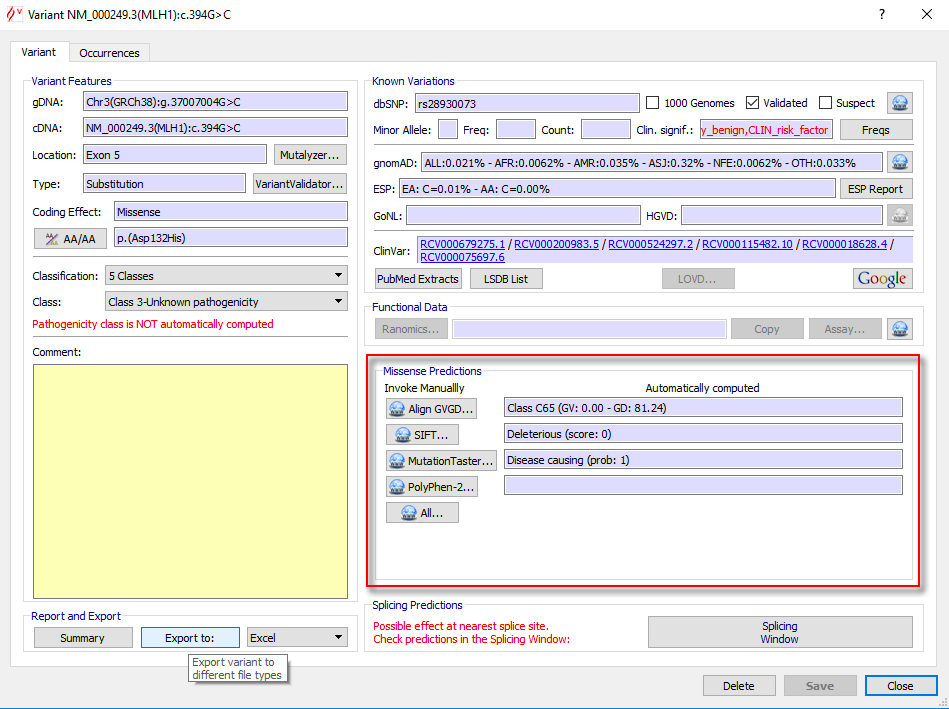
This section describes how automatically computed missense predictions are provided in Alamut® Visual.
Align GVGD predictions are computed using the orthologue alignment provided by Alamut® Visual for the gene under study. See also Orthologue Alignments.
Align GVGD scores interpretation: here.
Current version for precomputed predictions: Align GVGD Tavtigian et al. (2006)
SIFT predictions are computed using the orthologue alignment provided by Alamut® Visual for the gene under study. See SIFT Aligned Sequences.
SIFT scores interpretation: here.
Current version for precomputed predictions: SIFT 6.2.0
MutationTaster predictions are computed using the mapping between NCBI RefSeq and Ensembl Transcript Ids available in Alamut® Visual.
MutationTaster scores interpretation: here.
![]() MutationTaster
automatic predictions can be activated or deactivated with the Options dialog box
(menu 'Tools' > 'Options' > 'Display' tab).
MutationTaster
automatic predictions can be activated or deactivated with the Options dialog box
(menu 'Tools' > 'Options' > 'Display' tab).
Current version for precomputed predictions: MutationTaster2 from MutationTaster website
The PolyPhen-2 automatic predictions are not available due to licencing reasons (March 2013).
You can query the prediction site with an automatically filled web form by clicking on "PolyPhen-2..." button.
NGRL Manchester Multiple sequence alignment use in missense prediction tools
Tavtigian SV, Deffenbaugh AM, Yin L, Judkins T, Scholl T, Samollow PB, de Silva D, Zharkikh A, Thomas A. Comprehensive statistical study of 452 BRCA1 missense substitutions with classification of eight recurrent substitutions as neutral. J Med Genet. 2005 Jul 13
Ng PC, Henikoff S. Predicting Deleterious Amino Acid Substitutions. Genome Res. 2001 May;11(5):863-74. PubMed
Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, Kondrashov AS, Sunyaev SR. A method and server for predicting damaging missense variants. Nat Methods 7(4):248-249 (2010)
Schwarz JM, R�delsperger C, Schuelke M, Seelow D. MutationTaster evaluates disease-causing potential of sequence alterations. Nat Methods. 2010 Aug;7(8):575-6.
Special thanks to the Genetic Cancer Susceptibility Group at IARC for their kind help in implementing the Align GVGD algorithm.
© 2020 Interactive Biosoftware - Last modified: 14 may 2020